Scientists at EPFL have developed a technique that can be a game-changer for genetics by making the characterization of DNA-binding proteins much faster, more accurate, and efficient.
Genes hold the DNA code for producing all the proteins of the cell. To begin this process, genes require a huge family of DNA-binding proteins called transcription factors, which are of enormous interest to biologists today. However, we still know very little about many transcription factors because of their large number, their ability to combine into different pairs, and the difficulty of studying their DNA-binding properties in the lab. Now, EPFL scientists have developed a microfluidics-based technique that can greatly speed up the process, with only a minimum of materials. Publishing in Nature Methods, the researchers have already used it to determine the DNA-binding properties of over 60 transcription factors, including nine new ones.
Transcription factors
Mammals — including humans — have between 1300-2000 transcription factors, many of which combine with others into “heterodimers” in order to bind genes and induce their transcription into RNA. Heterodimers are estimated to range between 3000 and 25,000.
Consequently, the number of possible combinations can be very high. But we also need to understand their DNA-binding properties, e.g. their affinity and specificity for DNA. This is key if we are to ever exploit transcription factors for biotechnological or pharmaceutical purposes in the future. But profiling transcription factors is a daunting and very complex task, since it requires relatively large amounts of hard-to-make transcription factors.
There are several databases available, but in total they cover only around 500 single transcription factors, and only a fraction of heterodimers. Progress in this field is slow, and the available profiles in these databases are not very good in predicting which genes these transcription factors work on.
A microfluidics approach
The lab of Bart Deplancke at EPFL’s Institute of Bioengineering has now invented a new technique called SMiLE-seq, which can greatly speed up the process with only tiny amounts of transcription factors. The technique uses microfluidics, which control tiny amounts of liquids in equally tiny spaces. Microfluidics is fast-becoming an area of excellence at EPFL, bringing together a number of different fields and disciplines.
SMiLE-seq works by attaching small amounts of the transcription factor (or factors when studying heterodimers) in a microfluidic device — this is a chip with micrometer-size channels that allow liquid to flow through. Once the transcription factors attach to the chip’s surface, a large library of random DNA is pumped into the chip and flows over them. This allows the transcription factors to recognize their corresponding DNA sequences. The transcription factor-DNA complex is then physically trapped, while the DNA that is not bound is washed away.
Next, the bound DNA is taken off the device and prepared for sequencing to identify which part of it got caught by the transcription factors. This information is fed into specialized software that works out the DNA-binding properties of the transcription factors or heterodimers. In turn, this helps to better predict their DNA-binding profiles in living cells.
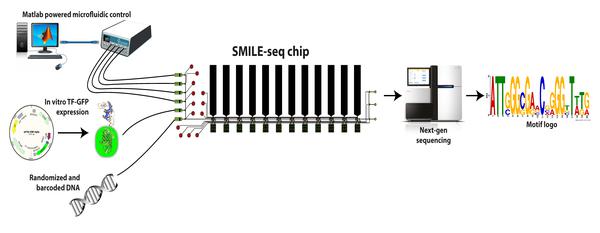
SMiLE-seq offers three advantages: First, it cuts down on the amount of transcription factors, as it only needs picograms of them. Second, it speeds up the process from days to less than an hour. And finally, SMiLE-seq is not limited by neither the length of the DNA target sequence, nor is it biased toward stronger affinity protein-DNA interactions.
Deplancke’s team used SMiLE-seq on 67 full-length human, mouse, and Drosophila transcription factors, successfully analyzing several that have never been studied before. He next plans to exploit the technique’s versatility for other molecules, e.g. RNA. His team has filed a patent, and a startup will take the concept of SMiLE-seq into the commercial world.
This work involved a collaboration of EPFL’s Institute of Bionegineering with its Swiss Institute of Experimental Cancer Research (EPFL-ISREC), Global Health Institute (EPFL-GHI) and the Swiss Institute of Bionformatics. It was funded by the Swiss National Science Foundation, SystemsX.ch, and EPFL.
Reference
Alina Isakova, Romain Groux, Michael Imbeault, Pernille Rainer, Daniel Alpern, Riccardo Dainese, Giovanna Ambrosini, Didier Trono, Philipp Bucher, Bart Deplancke. SMiLE -seq identifies binding motifs of single and dimeric transcription factors.Nature Methods 16 January 2017. DOI: 10.1038/nmeth.4143
Nik PapageorgiouMediacom